
Antiviral Activity
in Silico of Benzylisoquinoline Alkaloids
existing in
Berberis lilloana and B. commutata against
key Proteins of SARS-Cov-2
Actividad Antiviral in Silico de Alcaloides Bencilisoquinolinas Presentes en
Berberis lilloana y B. commutata Contra Proteínas Claves Del SARS-Cov-2
María A. Zígolo
Instituto de
Investigaciones para la Industria Química (INIQUI), Consejo Nacional de
Investigaciones Científicas y Técni- cas (CONICET).
Facultad de Ciencias Naturales,
Universidad Nacional de Salta, Av.
Bolivia 5150, (4400) Salta, Argentina.
maz130685@gmail.com
Abstract
SARS-CoV-2, a new strain of coronavirus (CoV),
was identified in Wuhan, China, in 2019, and has been threatening public health worldwide.
The aim of this work was to evaluate protoberberine alkaloids compounds of vegetal
origin as potential
SARS-CoV-2 inhibitors through docking studies.
Three key proteins of SARS-CoV-2, recently crystallized, were used as molecular targets: the spike glycoprotein (S), the main protease (Mpro) and the RNA-dependent RNA polymerase (RdRp).
Molecular docking was performed
using AutoDock, with the Lamarckian Genetic Algorithm, to analyse the probability
of docking. The best energy binding values for S protein were, in kcal/mol: -10.67 for Jatrorrhizine,
-9.65 for berberine, -9.22 for 5, 6-dihydroconstrictosine. For Mpro, they were, in kcal/mol: -10.15 for 5,6-dihydroconstrictosine, -9.86 for jatrorrhizine, -8.48 for berberubine. Finally, the best binding values for RdRp were, in kcal/mol: -9.04 for belambine,
-8.99 for canadine and -8.90 for berberine.
Key hydrogen bonds
and hydrophobic interactions between protoberberine alkaloids and the respective viral proteins were identified. These results suggest that these alkaloids could potentially be useful as drugs to be experimentally evaluated
against COVID-19.
Keywords: Binding affinity; Docking;
Protoberberine alkaloids; SARS-CoV-2
Resumen
El SARS-CoV-2, una nueva cepa de coronavirus identificada en Wuhan, China, en 2019, ha estado amenazando la salud pública en todo el mundo. El objetivo de este trabajo fue estudiar compuestos alcaloides de protoberberina de origen vegetal
como potenciales inhibidores del SARS-CoV-2
mediante estudios de acoplamiento. Se emplearon como dianas moleculares tres proteínas claves del SARS- CoV-2, recientemente cristalizadas: la glicoproteína de pico (S), la proteasa
principal (Mpro) y la ARN polimerasa dependiente de ARN (RdRp).
El acoplamiento molecular
se realizó mediante
AutoDock, con el Algoritmo Genético Lamarckiano, para analizar la probabilidad de acoplamiento. Los mejores
valores de energías de unión para la proteína S fueron, en kcal / mol: -10,67 para jatrorrizina, -9,65 para berberina
y -9,22 para 5,6-dihidroconstrictosina. Para Mpro fueron,
en kcal/mol: -10,15
para 5,6-dihidroconstrictosina, -9,86 para jatrorrizina y -8,48 para berberubina. Para RdRp fueron, en kcal/mol:
-9.04 para berlambina, -8.99 para canadina,
y -8.90 para berberina. Se identificaron enlaces de
hidrógeno claves e interacciones hidrofóbicas entre algunos de los alcaloides de protoberberina y las
respectivas proteínas virales.
De estos resultados surge que estos alcaloides podrían ser potencialmente útiles como fármacos antivirales a ser evaluados
experimentalmente contra COVID-19.
Palabras clave: Afinidad de unión; Alcaloides de protoberberina; Docking;
SARS-CoV-2
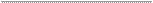
Zigolo, M. A. (2024). In Silico Antiviral Activity from Berberis lilloana and B. commutata. Protoberberine Alcaloids Against Key Proteins SARS-Cov-2. Revista Ciencias Naturales, 2(1), 10–21.
Recibido: 26/7/2023 Aceptado: 21/2/2024 Publicado: 15/3/2024 Editora:
María Victoria García
INTRODUCTION
In 2019, a new coronavirus caused an
outbreak of pulmonary disease in the city of
Wuhan, the capital of Hubei province
in China, and has since spread globally
and represent a challenge to public health. This virus has been named SARS-CoV-2, because the RNA genome is about 82% identical to the SARS coronavirus (SARS-CoV). The disease
caused by SARS-CoV-2 is called COVID-19 (Wu et al.,
2020; Zhou et
al., 2020). Human-to-human transmission is very fast.
(Li, 2020). However, waiting
for a vaccine, there are currently no effective medications against 2019-nCoV/ SARS-CoV-2.There is an urgent need
for the development of effective
prevention and treatment strategies for COVID-19 disease. Taking into account that although many of the vaccines developed promise to be very
effective, the period
of immunity they will
generate in the population is unknown. (Detoc et al., 2020; Kaur & Gupta,
2020; Malika et al., 2020).
Many drugs known for their effectiveness against other viruses are being investigated in search of a solution to the rapid and lethal advance of the new SARS-CoV-2. Some protease inhibitors used for treatment of patients with Human
Immunodeficiency Virus infection and acquired immune
deficiency syndrome (HIV/AIDS) or neuraminidase
inhibitors or nucleotide analogues like Remdesivir (GS-5734™) used for other pa- thogenic CoVs (Khaerunnisa et al., 2020).
Too
the chloroquine/hydroxychloroquine extensively used against malaria
and pre- viously described as
a potent inhibitor of most coronaviruses in vitro, including SARS-CoV-19 (White et al., 2014; Devaux et al., 2020). Plants
are great producers of secondary metabolites with biological activity
very interesting (Tallei
et al., 2020; Yang et al., 2018). The antiviral activitys of Many of these secundary metabolites have been described in numerous medicinal plants and
associated with compounds
like flavonoids, heterosides, terpenes,
and triterpenes, organic acids,
alkaloids, saponins, and quaternary ammonium
salts, among others (Roy & Saraf, 2006; Cecil et al., 2011; Wang et al., 2017; Khaerunnisa et al., 2020).
Many species containing protoberberin alkaloids have long been used in traditional
medicine in India, China, Tibet, and Japan primarily as antimicrobial remedies (Leitao Da-Cunha et
al., 2005). The antiviral activity of berberine-related alkaloids
was demonstrated and many of the infective mechanisms have been established. In respiratory syncytial
virus infection (RSV), (Shin et al., 2015) in several strains
of influenza A H1N1 (Cecil et al.,
2011; Enkhtaivan et al., 2018) and in the hepatitis C virus
(Hung et al.,
2018).
Docking tools give information from protein-ligand interactions. This is critical when rushing in the search of metabolites that can
cure or ease symptoms of diseases when time
and resources for extensive experimental work are scarce. Recently, in the last months, many SARS-CoV-2
macromolecules have been crystallized which has been repositioned
in the Protein Data Bank (PDB) and is
accessible by the public. This allow more effective simulations may be performed than if
model proteins from other closely
related viruses have to be used (Jin et al., 2020; Wrapp et al., 2020; Zhang et al., 2020). One of the best characterized drug targets
among coronaviruses is the main protease (Mpro, also called 3CLpro) (Anand
et al., 2003; Joshi
et al., 2020, 2021).
This enzyme is essential for processing the polyproteins that are translated from the viral RNA (Hilgenfeld, 2014). Cleavage by 3CLpro
and others viral proteases occurs
at a conserved glutamine catalitic
residue via the protease CYS-HIS dyad in which the cysteine thiol functions as the nucleophile in the proteolytic process (Anand et al.,
2003). In the initial
stage of the SARS-CoV-2 replication cycle, the attachment of the virion to the host cell is
initiated by interactions between
the S protein (spike) and human
angiotensin-converting enzyme (ACE2 receptor)
(Hoffmann et al., 2020). The S protein-receptor interaction is the primary
determinant for a coronavirus
to infect a host species and governs the tissue tropism of the virus. Structurally, the coronavirus spicule
is a trimer arranged asymmetrically. Each monomeric unit has three segments: an extensive ectodomain, a single-pass transmembrane segment, and a small
intracellular tail (Walls
et al., 2016).
The ectodomain contains
two subunits; the S1
subunit allows binding to the ACE2 receptor while the S2 allows
the fusion of viral and host
membranes. Two major domains
in S1, N-terminal domain (S1-NTD) and C-terminal domain (S1-CTD), have been identified.
The S1-CTD, also known as the receptor- binding
domain (RBD), is responsible for recognizing protein receptors ACE2 (Liu et al., 2015). Also, the spike protein
exists in two structurally distinct conformations, prefusion and postfusion.
All RNA viruses encode an RNA- dependent RNA polymerase (RdRp) that catalyzes the synthesis of their RNAs. The
polymerase RdRp is a crucial viral enzyme in
the life cycle of RNA viruses; due this, it has
been targeted in various
viral infections, who the hepatitis
C virus (HCV), the Zika virus
(ZIKV), and coronaviruses (CoVs) (Ganesan & Barakat, 2017; Elfiky et al., 2018, 2019).
The RdRp active site is highly conserved, with two successive and surface-accessible
aspartates in a beta-turn structure (Doublie & Ellenberger, 1998).
This protein is an important target for new antiviral drugs for SARS-CoV2.
In the present study, we investigated commun
protoberberine alcaloids from various vegetal species
as potential inhibitor
candidates for COVID-19
protease Mpro, the glycoprotein S and the RNA polymerase RdRp. These proteins are essential to the
transmission and virulence of the virus.
By inhibiting anyone of these proteins
or all, for a higher active
therapy, the severity of the infection will be reduced.
Our efforts have been placed in competitively inhibiting the binding of its natural substrates. The findings of the present study will provide other
researchers information to identify the right
drug to combat COVID-19.
MATERIALS AND METHODS
Viral proteins
Main protease (Mpro)
SARS-CoV-2 Mpro was crystallized in a complex
with N3 inhibitor and coordinates and structure factors were deposited in Protein
Data Bank (PDB ID: 6LU7) (Jin et al., 2020). The main protease monomer
contains three domains. Domains
I and II (residues 8-101 and 102-184) are made of antiparallel-barrel structures
in a chymotrypsin-like fold
responsible for catalysis. Binding pocket with
the main residues: THR24, THR26,
HIS41, PHE140, ASN142, GLY143, CYS145,
HIS163, HIS164, GLU166, and
HIS172. Contain the CYS-HIS catalitic
dyad.
S protein
Due to the crucial
importance of SARS- CoV-2- S spike protein,
the characterization at the prefusion
structure has been deposited
in the Protein Data Bank (PDB ID: 6VSB)
(Wrapp et al., 2020).
RNA polymerase (RdRp)
RNA polymerase (RdRp) bound to its
essential co-factors, nsp7 and nsp8 was
crystallized and coordinates and structure factors
were deposited in Protein Data Bank (PDB ID: 6NUR)
(Kirchdoerfer & Ward, 2019).
Antiviral compounds
Different protoberberine alkaloids, of plant
origin, with known antimicrobial activity, were
selected and evaluated
for their interaction with the two viral proteins.
In addition, synthetic
drugs at the present time used for the
treatment of COVID-19 were used as controls. The alkaloids three-dimensional structure were obtained
from database PubChem (https://pubchem.ncbi.nlm.nih.gov/): canadine (CID 21171), artavenustine (CID 181939), berberubine(CID 72704), berlambine (CID 11066), berberine (CID 2353), jatrorrhizine (CID 72323), lambertine (CID 10217), protopine (CID 4970), cryptopine (CID 72616),
quinolizinium, 2,10 dihydroxy-13-oxidodibenzo (CID 148262), constrictosine (CID 10016432), pallimamine (CID 132542920 ), orientalidine (CID 185550),
5,6-dihydroconstrictosine (CID 10401464). The synthetic nelfi (CID 64143)
and atazanavir (CID 148192), were used as controls for the interaction with Mpro and acyclovir (CID 135398513) and emtricitabine (CID 60877) to RdRp protein and and umifenovir (CID 131411) to S protein.
The ivermectin (CID 9812710), chloroquine (CID 2719), hydroxychloroquine (CID 3652), were used like a control
for the three proteins.
Docking analysis
Docking calculations were carried out with AutoDock
software (consist of two generations of software: AutoDock 4 and AutoDock
Vina). This software uses Lamarckian genetic algorithm (LGA) for calculations (Morris et al.,
2009). Number of genetic algorithm (GA) runs was set to 200 for each case analyzed. The Autodock 4 program was applied considering
all rotatable bonds for ligands and the whole
protein as a rigid structure. For the location and extent of the 3D area the search
space was defined by specifying a center,
the number of points at each dimension
and the points between spaces to focus the search space in the enzyme active site or putative binding
region.
For Mpro, the grid box center coordinates corresponded to the C atom of the histidine
residue 163: x (-19.055), y (16.637),
and z (64.100).
Dimensions and extention of the grid box were 90 x 90 x 90 and point spacing was
0.492 Å. For the S protein,
the grid box center coordinates corresponded to the oxygen atom of the alanine 419 residue for chain B: x (232.020), y (245.760), and z (265.659). Dimensions of the grid box were 124 x 124 x 124 and point spacing was 0.869 Å. Finally for the RNA polymerase RdRp, the grid box center coordinates corresponded to the nitrogen atom of the lysine 533 residue: x (142.000), y (139.000)
and z (150.000). Dimensions of the grid box were 114x 114x 114 and point spacing was 0.469Å.
Prior to docking
Mpro, the inhibitor N3 and water molecules were removed from the protein
structure. Polar hydrogen
atoms were added, and Gasteiger atom charges
were assigned to protein atoms. Other parameters were set to default values.
The 200 conformers found for each compound with the Autodock program were grouped
in clusters that were ordered according to a ranking,
which was determined by the stability of the enzyme-ligand complexes within each cluster. Clustering of the 200 conformers was done according to the similarity with the conformation adopted inside the enzyme.
The criterion used to
evaluate such similarity was the residue mean quadratic square root deviation (RMSD). The residues
were obtained through the difference between the atom coordinates of a given conformer respect to the cluster to which the most
stable conformer belongs.
Each cluster grouped conformers
with RMSD lower or
equal to 2.0 Å.
For visualization of protein-ligand complexes the software Visual Molecular Dynamics 1.9.1 (VMD) was used (Theoretical and Computational Biophysics Group, University of Illinois).
RESULTS
Selection of conformers
From the 200 docking
runs for all the
evaluated compounds, both from natural origin and the synthetic
controls, the binding energies for each conformer
were obtained which,
in turn, were grouped into clusters.
Each compound was represented by the most populated clusters, which in some cases were
also those with the lowest binding energies. The binding energies for the interactions of each compound with the corresponding
protein are shown in Table 1.
When Mpro protein interacted with natural
compounds, similar values
than those observed with the synthetic antivirals atazanavir and nelfinavir, were found. The strongest affinities for 5,6-dihydroconstrictosine and Jatrorrhizine, with ΔG of -10.15 and -9.86
kcal/mol, were obtained, respectively. Other
|
Alcaloid
|
Skeleton type
|
Mpro ΔG
(kcal/mol)
|
S Protein
ΔG
(kcal/mol)
|
RpRp ΔG
(kcal/mol)
|
|
Canadine
|
I
|
-7.69
|
-7.36
|
-8.99
|
|
Artavenustine
|
I
|
-7.33
|
-7.36
|
-7.76
|
|
Berberubine
|
II
|
-8.48
|
-7.67
|
-8.89
|
|
Berlambine
|
II
|
-7.38
|
-7.12
|
-9.04
|
|
Berberine
|
III
|
-7.35
|
-9.65
|
-8.90
|
|
Jatrorrhizine
|
III
|
-9.86
|
-10.67
|
-7.57
|
|
Lambertine
|
IV
|
-7.82
|
-9.20
|
-8.65
|
|
Protopine
|
V
|
-7.66
|
-7.40
|
-8.04
|
|
Criptopine
|
V
|
-7.94
|
-7.92
|
-7.80
|
|
Constrictosine
|
VII
|
-7.19
|
-7.20
|
-8.75
|
|
Pallimamine
|
VIII
|
-7.41
|
-9.11
|
-8.35
|
|
Orientalidine
|
IX
|
-7.36
|
-8.03
|
-7.41
|
|
5,6-dihydroconstrictosine
|
X
|
-10.15
|
-9.22
|
-7.57
|
|
Atazanavir
|
|
-9.47
|
-
|
|
|
Nelfinavir
|
|
-8.25
|
-
|
|
|
Umifenovir
|
|
-
|
-7.47
|
|
|
Acyclovir
|
|
-
|
-
|
-5.88
|
|
Emtricitabine
|
|
-
|
-
|
-4.60
|
|
Chloroquine
|
|
-5.11
|
-7.25
|
-4.10
|
|
Hydroxychloroquine
|
|
-4.20
|
- 5.30
|
-5.10
|
|
Ivermectine
|
|
-6.70
|
-8.71
|
-6.03
|
Table 1. Gibbs free energy (ΔG, in kcal/mol)
calculated for the interaction between
SARS-CoV-2 and different molecular
targets Mpro, S protein, RNA polymerase RdRp and selected
alkaloids. Synthetic antivirals used as controls are in bold.
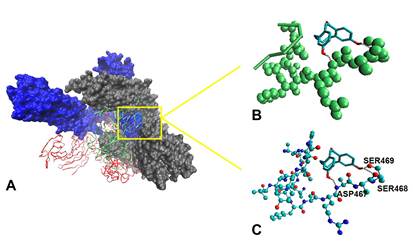
Figure 1. Three-dimensional (3D) molecular interaction of 5, 6 -dihydroconstrictosine with S glycoprotein.
A. Structure of spike protein S: chain A (blue), chain B (red) and chain C (grey). Domain RBD of chain B (green ribbons). B.
Residues 420 to 425 from domain RBD of chain B (green trace) and residues 460 to
461 (green spheres). The 5, 6- dihydroconstrictosine is represented in licorice form (cyan carbon atoms,
compounds showed interesting affinities for Mpro, for example:
berberubine, canadine and cryptopine (Table 1). On the other hand,
the plant-derived natural
compounds evaluated for S
protein, yielded very
high affinity and
slightly highervaluescomparedtocontrols.
Thehighest affinities were observed for jatrorrhizine,
berberine, and 5,6-dihydroconstrictosine. The
most interesting results
were observed for jatrorrhizine and berberine (-10.67and -9.65 kcal/mol, respectively), which presented more affinity for the viral S protein
compared to the synthetic antivirals umifenovir, ivermectin, chloroquine, and hydroxychloroquine (-7.47, -8.71, -7.25, and -5.30 kcal/mol,
respectively) (Table 1).
Analysis of the molecular interactions of candidate
compounds with the proteins
evaluated.
The 5, 6- dihydroconstrictosine, berberine and jatrorrhizine showed the best interactions with the spike protein
with the most favorable
Gibbs free energies (-9.22, 9.65 and
10.67 kcal/mol respectively) (Table 1). 5, 6- dihydroconstrictosine interacted with a great number of residues from RBD domain
(Fig. 1A). Whit residues
420 to 425 and 460 to
463 of chain B (Fig. 1B). There was a total of three hydrogen bonds with residues
ASP467, SER469 and SER468 of the RBD domain of B chain (Fig. 1C).
Key interactions of possible
candidate drugs with Mpro were analyzed:
5, 6- dihydroconstrictosine, jatrorrhizine, berberu-
bine and nelfinavir (Fig. 2).
All the natural and semisynthetic active compounds were bounded to the enzyme active site, as well as the controls nelfinavir and atazanavir. The 5, 6-dihydroconstrictosine presented five hydrogen bond interactions
(Fig. 2A): two with THR26 (2.51 Å and 2.73 Å),
one whit GLY143 (1.33 Å), one with ASN142
(2.01 Å) and one with CYS145
(1.02 Å) implicated in CYS-HIS
dyad. It also showed hydrophobic interactions with other residues from catalytic site.
All these interactions increased the binding affinity
with respect to the control compound
nelfinavir, which presented only one hydrogen
bond interactions with GLU166 (2.33
Å) and
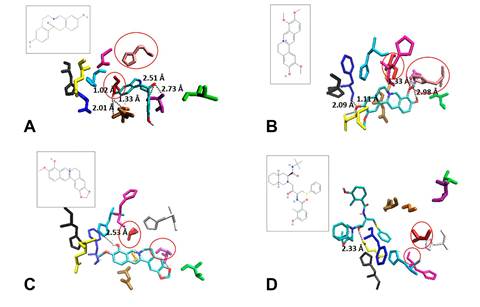
Figure 2. Three-dimensional (3D) molecular interaction of compounds: A.
5,6-dihydroconstrictosine. B. Jatrorrhizine. C. Berberubine. D. Nelfinavir with Mpro. Active
site residues are represented with sticks: THR24 (green), THR26 (purple), HIS41 (pink), PHE140 (blue), ASN142 (brown), GLY143 (orange),
CYS145 (red), HIS163 (cyan), HIS164 (magenta), GLU166 (yellow), and HIS172 (black).
The black dotted lines, indicated Hydrogen bond. The residues CYS145 and HIS41 are
indicated by red circle.
hydrophobic
interactions with other
protease catalytic residues (Fig. 2D). Jatrorrhizine, had four hydrogen bond interactions with catalytic residues HIS41 (implicad
in CYS- HIS dyad),
HIS164, GLU166 and PHE140
(2.98 Å, 1.33 Å, 2.09 and 1.11 Å respectively) and hydrophobic interactions with the other catalytic residues (Fig. 2B).
Berberubine interacted with the Mpro mainlybyhydrophobicinteractions, presenting only one hydrogen bond interaction with the
HIS166 catalytic residue (Fig. 2C).
Jatrorrhizine and berberine
interacted with residues 350 to 460 from RBD domain of B
chain and the some residues
of chain C. In
contrast the control drug nelfinavir interacted only with residues of chain C (Fig. 3).
Berlambine, canadine
and berberina
binding to RdRp in NSP12, chain A (blue).
All three alkaloids interact with the NSP12 chain through hydrophobic interactions with various residues of the polymerase including the ASP255 and ASP256 active site residues (Fig. 4).
DISCUSSION
Plants have a long
evolutionary history of developing resistance against viruses. The ability to produce secondary
metabolites, has generated a wide range
of possible sources
for antiviral drugs. Medicinal plants have been used
ancestrally in different civilizations. The history have showed their low toxicity and their minimal
adverse effects at the doses used. In this sense, representing an important advantage as compared with new synthetic
drugs, which need to be evaluated for safety
through strict clinical trials (Abbas et al., 2016; Horden, 2016).
Phytocomplexes are mixtures of active
compounds, formed by isomers and analogue
compounds, with the capacity for synergistic activity and contains compounds that help
mitigate the toxicity of the main active drugs.
This help microorganisms are less likely
to develop resistance
to a phytocomplex than to a unique compound
in a synthetic drug
(Alonso, 2008; Bruneton, 2001; Camponovo & Bandoni,
1995). In this study, we analyzed
variosus active compounds that are commonly found in the alkaloid
extracts of various
species of the Berberidaceae family and that have been used throughout history as medicinal
plants.
It is
important to highlight that all
the
evaluated protoberberine alkaloids bound with good affi to the three evaluated SARS- CoV-2 targets. Therefore, we believe that they could act through diff mechanisms of action.
The alkaloids 5, 6- dihydroconstrictosine and jatrorrhizine
interact with key residues
in main protease Mpro involved in substrate
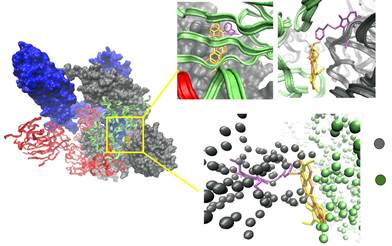
Figure 3. Three-dimensional (3D) molecular interaction of the compounds: jatrorrhizine (yellow), berberine
(orange) and umifenovir
(purple) with S glycoprotein. Structure
of spike protein S: chain A (blue), chain B (red)
and chain C (grey). Domain
RBD of chain B (green
ribbons). Residues 150 to 157 and 176 to 200 from
chain C (grey spheres). RBD residues 350 to 460 from chain B (green spheres).
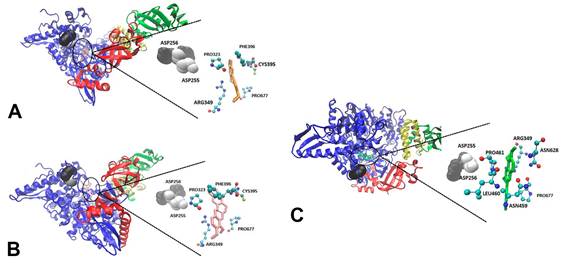
Figure 4. Three-dimensional (3D) molecular interaction of berlambine (A), canadine (B) and berberine (C)
with polymerase RdRp.
Black circles indicate
the RdRp site where ligands
bind. The two active site residues
are shown as black (ASP256) and white (ASP255) spherical surface.
binding, including the CYS145-HIS41 dyad. This is an important factor for viral
proteins sintesys inhibition. This factor was considered for the synthesis
of potential inhibitors of the main protease Mpro structure-based ligand design. Macchiagodena et
al. (2020), obtained a gibb-free energy value of -8.92 kcal/mol
from to the most potent ligand for 3CLpro
protease, which linked of the dyad CYS-HIS (Macchiagodena et
al., 2020). The compounds 5, 6-dihydroconstrictosine and jatrorrhizine
showed higher free Gibbs energy (-10.15
and
-9.61 kcal/mol).
The most studied
protoberberine alkaloids is a Berberine. Berberine is marketed in various countries around the world as dietary supplements
(BIOTICS RESEARCH®, DR. MEROLA®). It has antimalarial, antibacterial, and antiviral effects,
among other biological activities (Wang et al., 2017).
Berberine can also inhibit the replication of influenza A in vitro in several
different cell types and with two influenza A (H1N1) strains (Cecil et al., 2011). As in this study,
other researchers found that berberine
displayed binding energy of -7.91
kcal/mol to Mpro (in this work -7.35 kcal/mol
to Mpro and -9.65 kcal/mol to glycoprotein S) and had the
ability to producing conformational changes in the target enzyme. (Agrawal
et al., 2020). In this work we show that it also binds to the
spike protein S (-9.65 kcal / mol) and to the
RdRp polymerase (-8.90) with higher affinity than in the Mpro protease.
This makes it a
very interesting compound
to be clinically evaluated in COVID-19 considering that it is already
commercially available and there are several laboratories that manufacture
it.
Hydrophobic
interactions played a particularly important role for the Mpro protein. In the case of berberubine and nelfinavir, which showed a good binding
energy value, showed one only hydrogen
bond and interactions of hydrophobic nature
with the protease
residues. Khaerunnisa et al. (2020) also showed the importance of hydrophobic
interactions for a series of compounds of plant
origin in the interaction with the viral
protease Mpro (Malika et
al., 2020).
All of
the compounds
evaluated against S glycoprotein showed very favorable binding energies,
higher than those for the
synthetic antiviral compounds,
umifenovir (-7.47 kcal/mol), chloroquine (-7.25 kcal/mol), hydroxychloroquine (-5.30 kcal/mol) and ivermectin (-8.71 kcal/ mol) directed to that viral target. The most favorable binding energies were obtained for jatrorrhizine (-10.67), berberine (-9.65kcal/mol), 5, 6-dihydroconstrictosine (-9.22 kcal/mol), lambertine (-9.20 kcal/mol) and pallimamine (-9.11 kcal/mol). These results are very promising. Others
works, where the coupling of natural
and synthetic compounds with viral
glycoprotein S, were analyzed showed
best binding energies
obtained for S protein were:
-11.55 kcal/mol to coenzyme
A, -11.089 kcal/ mol to flavin adenine dinucleotide, and - 9.36 kcal/mol to tiludronate (Hall &
Ji, 2020).
In agreement with the low Gibbs free energy, for the 5, 6- dihydroconstrictosine, jatrorrhizine and brberine numerous
molecular interactions were observed, all of them with
the RBD region (recognition of ACE human receptors), chain B, were observed. Therefore, the three alkaloids would be a good candidate to be evaluated experimentally to
learn how it would affect the glycoprotein activity.
The5,6-dihydroconstrictosineisaarotopine alkaloids.
This group of protoberberine
alkaloids are apparently not very common.
Their occurrence in Aristolochiaceae family was strictly confined to A. constricta. Protopine alkaloids are usually reported as C-2 substituted on the basis of biogenetic
considerations. The unusual
absence of C-2 substitution was noticed in the protopines of Aristolochia family (Rastrelli et al., 1997).
Jatrorrhizine is found in some plant species from the families Papaveraceae, Berberidaceae, Menispermaceae, Ranunculaceae, and Ruta- ceae (Cecil et al., 2011). This compound
has demonstrated inhibits mammary carci- noma cells (Sun
et al., 2019), in
vitro and in vivo antitumor
activities (Qin et al., 2019),
antihypercholesterolemic eff (Wu et al., 2014). 5,6-dihydroconstrictosine, Jatrorrhi-zine and berberine interacts whit many hydrophobic interactions with the spike protein residues. It also formed hydrogen bonds. Many of these interactions of diff nature were established
in the RBD region of S glycoprotein. For this reason, we consider that they three compounds are the most promising compound with potential to inhibit spike protein.
For RNA polymerase RdRp the best values
of binding energies were for Berlambine
-9.04 kcal /mol, Canadine -8.99
kcal / mol and berberine -8.90 kcal / mol. Other studies
evaluated the synthetic compounds rivavirin, tenofovir, sofosbuvir, IDX 184, setrobuvir and YAK as possible
inhibitors of RNA polymerase RdRp with binding energy values of (-7.8, -6.9,
-7.5, -9.0, -9.3 and - 8.4 kcal / mol respectively) (Elfiky, 2016) and azafluorene derivates (Venkateshan et al.,
2020).
The alkaloids with better binding
energies berlambine, canadine and berberine
binding to NSP12 in the active site (Fig. 4). This binding like and any union to its cofactors could affected the correct assembly of the
polymerase, preventing its correct functioning. For this we thinking
that berlambine, canadine and berberine would be good candidate
drugs to be experimentally evaluated against this SARS-CoV-2
polymerase.
In fact, being derived from medicinal
plants with ancestral use proven for oral consumption is an important
advantage of some natural
origin compounds. Should they
be administered as part of phytocomplexes, there might be a synergistic effect of the desired activity
while potential side effects of the main active
compounds, if they were given pure, would be attenuated. This important
advantage would facilitate the production and reduce the cost because fewer downstream operations would
be needed as there is no
need to obtain the high purity product.
(Cortés et al.,
2004; Sharapin et
al., 2000).
CONCLUSIONS
There are already several
drugs and va- ccines being assessed clinically
for SARS- CoV-2. However there is still a need to iden- tify additional treatments as alternatives tha vaccines, since the period of immunity that they generate in the population is unknown and also if we take into account
that there are people
who cannot be vaccinated for di-
fferent reasons. Identifying antiviral drugs from medicinal plants and sintetic drugs with side-effect profiles
known by clinicians are very important. In this sence, various proto- berberine alkaloids proposed in this work has been from known ancestral
uses and many of
them are sold as supplements dietary for oral consumption.
Based on the binding energies obtained
and that protoberberine alkaloids interact
with the key catalytic residues,
interaction conformations between the proposed natural compounds and protease Mpro, glycoprotein
S and
RNA polimerase RdRp proteins are highly
possible. In addition,
the tested natural compounds performed better than
the synthetic antivirals used as controls: nelfinavir and atazanavir, for protease Mpro, umifenovir for S protein
and acilovir and emtriacitabine for polymerase RdRp. Thus, the protoberberine
alkaloids could be potential active agents for treatment of COVID-19, either alone or as
adjunct therapies with other medications found to have antiviral
activity in vivo.
This study also contains valuable infor-
mation to increase
the knowledge of certain
protoberberine nucleus of compounds with best
affinities towards these molecular targets. Furthermore, the present study provides
molecular details that allow us to propose structural modifications of some compounds
to make the interaction between them and proteins even more effective. Everyday we hope to be better prepared and we know that
Effective antiviral treatments can save lives,
and we need to have them ready in the future.
ACKNOWLEDGMENTS
The author expresses gratitude to CONICET and acknowledges the contributions of the editor, reviewers, and Cintia Avila for image processing. This work was funded by CONICET.
REFERENCES
Abbas Zaidi, S., Jameel, S., Jafri, K., Khan, S., & Ahmad, E. (2016).
Ilaj bil hijamah (cupping
therapy) in the Unani system
of medicine:
anecdotal practice
to evidence-based therapy.
Acta Medico-Historica Adriatica, 14(1) 81-94.
Agrawal, A., Jain N. K., Kumar, N., &
Kulkarni, G.
 T. (2020). Molecular Docking Study to Iden- tify Potential
Inhibitor of COVID-19
Main Protease Enzyme: An In-Silico Approach. Chem Rxiv. https://doi.org/10.26434/chemr-xiv.12170904.v1
T. (2020). Molecular Docking Study to Iden- tify Potential
Inhibitor of COVID-19
Main Protease Enzyme: An In-Silico Approach. Chem Rxiv. https://doi.org/10.26434/chemr-xiv.12170904.v1
Alonso, J. (2008). Tratado de Fitofármacos y Nu- tracéuticos. (1° Ed). Reimpr.Corpus Edicio- nes, Rosario, Argentina.
Anand,
K., Ziebuhr, J, Wadhwani, P.,
Mesters,
J. R., & Hilgenfeld, R. (2003). Coronavirus main proteinase (3CLpro) structure:
Ba-
sis for design
of anti-SARS drugs.
Science, 300, 1763–1767. https://doi.org/10.1126/ science.1085658
Bruneton, J. (2001). Farmacognosia, fitoquímica, plantas medicinales. (2° Ed). Editorial ACRI- BIA S.A.,
Argentina.
Camponovo, L., & Bandoni, A. (1995). Farmaco- logía:
Materia Médica y Terapéutica. Buenos Aires: López Etchegoyen Libreros Editores, Argentina.
Cecil, C. E., Davis, J. M., Cech, N. B., & Laster,
S. M. (2011). Inhibition
of H1N1 influenza A virus growth and induction of inflammatory
mediators by the isoquinoline alkaloid
ber- berine and extracts
of goldenseal (Hydrastis canadensis). International Immunopharma- cology, 11(11),
1706-1714. https://doi.or- g/10.1016/j.intimp.2011.06.002
Cortés Gallardo,
V., Macedo Ceja, J., Hernández Arroyo,
M., Arteaga Aureoles,
G., Espinoza Galván, D., & Rodriguez Landa, J. (2004). Farmacognosia: breve historia de sus oríge- nes y su relación
con las ciencias médicas.
Revista Biomedica, 15, 123-136.
https://doi. org/10.32776/revbiomed.v15i2.381
Detoc, M., Bruel, S., Frappe,
P., Tardy, T., Botel- ho-Nevers, E., & Gagneux-Brunon, A. (2020). Intention to participate in a COVID-19
vacci- ne clinical trial and to get vaccinated
against COVID-19 in France during the pandemic. Vaccine, 38(45)
7002-7006. https://doi.or- g/10.1016/j.vaccine.2020.09.041
Devaux, C. A., Rolain, J. M., Colson,
P., & Raoult, D. (2020). New insights on the anti- viral effects of chloroquine against
corona- virus: what to expect for COVID-19. Inter- national
Journal of Antimicrobial Agents, 55(5):105938. https://doi.org/10.1016/j.ijan- timicag.2020.105938
Doublie, S., & Ellenberger, T. (1998).
The me- chanism of action of
T7 DNA polymera- se. Current
Opinion in Structural Biology, 8(6):704-12.
https://doi.org/10.1016/s0959-440x(98)80089-4
Elfiky, A. A., & Ismail A. (2019). Molecular dyna- mics and docking reveal the potency
of novel GTP derivatives against RNA dependent RNA polymerase of genotype 4a HCV. Life Scien- ces, 238, 116958. https://doi.org/10.1016/j. lfs.2019.116958
Elfiky, A. A., & Elshemey,
W. M. (2018). Mo- lecular dynamics
simulation revealed bin- ding of nucleotide inhibitors to ZIKV poly- merase over 444 nanoseconds, Journal of Medical Virology, 90(1), 13-18. https://doi. org/10.1002/jmv.24934
Elfiky, A. A., & Elshemey,
W. M. (2016). IDX- 184 is a superior
HCV direct-acting antivi-
ral drug: a QSAR study, Medicinal Chemis- try Research, 25(5):1005-1008. https://doi. org/10.1007/s00044-016-1533-y
Enkhtaivan, G., Kim, D. H., Park,
G. S., Pandu rangan, M., Nicholas, D.
A., Moon, S.H., Ka dam, A.A., Patel, R.V., Shin, H. S., & Mistry, B.M. (2018).
Berberine-piperazine conjuga- tes as potent
influenza neuraminidase blocker. International Journal of Biological Macro- molecules, 119, 1204-1210. https://doi.or- g/10.1016/j.ijbiomac.2018.08.047
Ganesan, A., & Barakat, K. (2017).
Applications of computer-aided approaches in the development of hepatitis C antiviral
agents, Expert Opin. Drug Discovery, 12(4), 407-425. https://doi.or g/10.1080/17460441.2017.1291628
Hall, D. C., & Ji, H. F. (2020).
A search for medi- cations to treat COVID-19
via in silico mo- lecular docking
models of the SARS-CoV-2
spike glycoprotein and 3CL protease. Travel Medicine
and Infectious Disease, 35, 101646. https://doi.org/10.1016/j.tmaid.2020.101646
 Hilgenfeld, R. (2014). From SARS to MERS:
Crystallographic studies on coronaviral
proteases enable antiviral drug design. The FEBS Journal., 281, 4085–4096. https://doi. org/10.1111/febs.12936
Hilgenfeld, R. (2014). From SARS to MERS:
Crystallographic studies on coronaviral
proteases enable antiviral drug design. The FEBS Journal., 281, 4085–4096. https://doi. org/10.1111/febs.12936
Hoffmann, M., Kleine-Weber, H., Krüger, N., Mü- ller, M., Drosten,
C., & Pöhlmann
(2020). The novel coronavirus 2019 (2019-nCoV) uses the
SARS-coronavirus receptor ACE2 and the ce- llular protease
TMPRSS2 for entry into target cells. BioRxiv.The preprint server
for biology. https://doi.org/10.1101/2020.01.31.929042
Horden, P. and Kukita Yoshikawa,
Naoë (2016). Me- dicine, Religion and Gender in Medieval
Cultu- re., 60(3), 441-442.
Hung, T.-C., Jasse, A., Liu, C.- H., Lin, Ch.-J., Lin,
Ch.- Ch., Wong, S. H., Wang, J. Y., Yen, M.- H.,
& Lin, L.- T. (2018).
Berberine inhibits hepatitis C virus entry by targeting
the viral E2 glyco- protein. Phytomedicine, 53, 62-69. https://doi. org/10.1016/j.phymed.2018.09.025
Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y.,
Zhang, B., Li, X., Zhang, L., Peng, C., Duan,Y.,Yu, J., Wang, L., Yang, K., Liu, F., Jiang, R., Yang, X., You, T., Liu, X., Yang, X., Bai, F., Liu, H., Liu, X., Guddat, L.W., Xu, W., Xiao, G., Qin, Ch., Shi, Z., Jiang, H.,Rao, X., & Yang, H., (2020). Struc-
ture of Mpro from SARS-CoV-2and discovery of its inhibitors. Nature.582, 289-293. https://doi. org/10.1038/s41586-020-2223-y.
Joshi, T., Joshi, T., Sharma, P., Mathpal, S., Pundir, H., Bhatt, V., & Chandra, S. (2020). In silico screening of natural compounds against CO- VID-19 by targeting Mpro and ACE2 using mole- cular docking. European Review for Medical
and Pharmacological Sciences, 24(8),
4529–4536. ht- tps://doi.org/10.26355/eurrev_202004_21036
Joshi, T., Bhat, S., Pundir, H., & Chandra, S. (2021). Identifi of Berbamine, Oxyacanthine and Rutin from Berberis asiatica as anti-SARS- CoV-2 compounds: An in silico study. Journal of Molecular Graphics & Modelling, 109, 108028. https://doi.org/10.1016/j.jmgm.2021.108028
Kaur, S. P., & Gupta, V. (2020). COVID-19 Vaccine: A comprehensive status report.
Virus
Research, 288, 198114.
https://doi.org/10.1016/j.virus- res.2020.198114.
Khaerunnisa, S., Kurniawan, H., Awaluddin, R., Su- hartati, S., & Soetjipto, S. (2020). Potential Inhi- bitor of COVID-19 Main Protease (Mpro) from Several Medicinal Plant Compounds by Mole- cular Docking Study. Preprints.org. https://doi. org/10.20944/preprints202003.0226.v1
Kirchdoerfer, R. N., & Ward, A. B. (2019). Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nature communica- tion, 10, 2342. https://doi.org/10.1038/s41467-
019-10280-3
Leitao Da-Cunha, E. V., Fechine, I. M., Guedes, D. N., Barbosa-Filho, J. M., & Sobral Da Silva, M. (2005). Protoberberine alkaloids. The Alka- loids, Vol. 62. https://doi.org/10.1016/S1099- 4831(05)62001-9
Liu, C., Tang, J., Ma,
Y., Liang, X.,
Yang Y, Peng, G.,
Qi, Q., Jiang, S., Li, J., Du, L., & Li, F. (2015). Receptor usage and cell entry of porcine epide- mic diarrhea coronavirus. Journal
of Virology,
89(11), 6121–6125. https://doi.org/10.1128%2FJ- VI.00430-15
Macchiagodena, M., Pagliali, M., & Procacci, P. (2020).
Identification of potential
binders of the main protease
3CLpro of the COVID-19
via structure-based ligand design and mole- cular modeling.
Chemical Physics Letters, 750, 137489.
https://doi.org/10.1016/j.cple- tt.2020.137489.
Malika, A. A., McFaddena, S. M., Elharakea, J., & Omera, S. B. (2020). Determinants of CO-
VID-19 vaccine acceptance in the US. ECli- nicalMedicine, 26, ID 100495.
https://doi. org/10.1016/j.eclinm.2020.100495
Morris, G. M., Huey, R., Lindstrom, W., Sanner,
M. F., Belew,
R. K., Goodsell, D. S., & Olson, A. J. (2009). Software
News and Updates. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor
Flexibility. Journal of Computational Chemistry, 30(16), 2785-2791. https://doi.org/10.1002/jcc.21256
Qin, Q.- P., Zou, B.-Q., Wang, Z.- F., Huang, X.-L., Zhang,
Y., Tan, M.- X., Wang, S.- L., & Liang, H. (2019). High in vitro and
in vivo antitumor activities of luminecent platinum (II) complexes with jatrorrhizine derivative. European Journal
of Medicinal Chemistry, 183, 111727.
https://doi.org/10.1016/j.ej- mech.2019.111727
Rastrelli, L., Capasso,
A., Pizza, C., De Tommasi, N., & Sorretino, L. (1997). New Protopine
and Benzyltetrahydroprotoberberine Alka- loids from Aristolochia constricta and Their Activity
on Isolated Guinea-Pig Ileum. Na- tural Products 60,11, 1065-1069.
https://doi. org/10.1021/np960710b
Roy, A., & Saraf, S. (2006). Limonoids: overview
of significant bioactive
triterpenes distributed in plants
kingdom. Biological and Pharma-
ceutical Bulletin, 29(2), 191-201.
Sharapin, N., Machado,
L., Souza, E., de Albuquer- que, E., Valverde, E., & López, J. M. (2000). Fundamentos de tecnología de productos fito- terapéuticos. Santa Fe de Bogotá, Convenio Andrés Bello y Red Iberoamericana de Pro-
ductos Fitofarmacéuticos (RIPROFITO) del subprograma X de
CYTED. Pp 248. España.
Shin, H.-B., Choi, M.-S., Yi, C.-M., Lee, J., Kim,
N.-J., & Inn, K.-S. (2015). Inhibition
of res- piratory syncytial
virus replication and vi- rus-induced p38 kinase activity by berbe- rine. International Immunopharmacology, 27, 65-68. http://dx.doi.org/10.1016/j.in- timp.2015.04.045
Sun, Y., Gao, X., Wu, P., Wink, M., Li, J., Dian, L., & Lian, Z. (2019). Jatrorrhizine inhi- bits mammary carcinoma
cells by targeting TNIK mediated Wnt/β-catenin signalling an
depithelial-mesenchymal transition (EMT). Pythomedicine, 63,153015. https://doi.or- g/10.1016/j.phymed.2019.153015
Tallei, T. E., Tumilaar, S. G., Niode, N. J., Fatimawa- li, Kepel, B. J., Idroes, R., Eff Y., Sakib, S.A., & Emran, T. B. (2020). Potential of Plant Bioactive
Compounds as SARS-CoV-2 Main Protease (Mpro) and Spike (S) Gly- coprotein Inhibitors: A Molecular Docking Study. Scientifica, ID 6307457. https://doi. org/10.1155/2020/6307457
Venkateshan, M., Muthu, M., Suresh, J., & Ranjith Kumar, R. (2020). Azafl derivatives as inhibitors of SARS CoV-2 RdRp: Synthesis, physicochemical, quantum chemical, modeling and molecular docking analysis. Journal of Mo-
lecular Structure,
1220, 128741. https://doi. org/10.1016/j.molstruc.2020.128741
Walls, A. C., Tortorici,
M. A., Bosch, B. J., Frenz, B., & Rottier, P. J. (2016). Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature, 531, 114–117. https://www.nature.com/articles/nature16988
Wang, W. J., Yang, T., Chen, H., Xu, Y. N., Yu, L.
F.,
Liu, T., Tang, J., Yi, Z., Yang, C.G., Xue, W., & Yang, F. (2017). The synthesis and an- tistaphylococcal activity
of 9, 13-disubtituted berberine derivatives. European Journal
of Medicinal Chemistry, 127,424-433. https:// doi.org/10.1016/j.ejmech.2017.01.012
White, N. J., Pukrittayakamee, S., Hien, T. T., Faiz, M. A., Mokuolu, O. A., & Dondorp, A. M.
(2014) Malaria. Lancet, 383:723–35.
Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C. L., Abiona, O., Graham, B. S.,
& McLellan, J. S. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion con- formation. Science, 367(6483), 1260-1263. http://dx.doi.org/10.1126/science.abb2507
Wu, F., Zhao, S., Yu, B., Chen, Y.-M., Wang, W., Song, Z.-G., Hu, Y., Tao, Z. W., Tian, J.-H., Pei, Y.-Y., Yuan, M.-L., Zhang, Y.-L., Dai, F.-H., Liu, Y.,Wang, Q.-M., Zheng, J.-J., Xu, L., Holmes, E. C., & Zhang, Y.-Z. (2020). A New coronavirus asso-
ciated with human respiratory
disease in China. Nature, 579, 265–269. https://doi.org/10.1038/ s41586-020-2008-3
Wu, H., Heb, K., Wang, Y. Z., Xue, D. F., Ning, N., Zou,
Z., Ye, X., Li, X., Wang, D. Z., & Pan,
J. (2014). The antihypercholesterolemic effect of jatrorrhizine is olated from Rhizoma Copti- dis. Phytomedicine. https://doi.org/10.1016/j.phymed.2014.05.002
Yang, L., Wen, K.S., Ruan, X., Zhao, Y. X., Wei, F., & Wang, Q. (2018). Response
of plant secondary metabolites to environmental factors. Molecules, 23(4), 762. https://doi. org/10.3390/molecules23040762
Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R.
(2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Nature. http://dx.doi. org/10.1038/s41586-020-2223
Zhou, P., Yang, X.L., Wang, X. G., Hu, B., Zhang,
L., Zhang, W., Si, H. R., Zhu, Y., Li, B., Huang, C. L., Chen, H. D., Chen, J., Luo, Y., Guo, H., Jiang, R. D., Liu, M. Q., Chen, Y., Shen, X. R., Wang, W., Zheng,
X. S., Zhao, K., Chen, Q. J., Deng, F., Liu, L. L., Yan, B.,
Zhan, F. X., Wang, Y. Y., Xiao, G. F., & Shi, Z. L. (2020).
A. Pneumonia outbreak
associated with a new coronavirus of probable bat origin. Nature, 579, 270–273.
https://doi. org/10.1038/s41586-020-2012-7